I’ve been working on a project with Quadry Chance (U. Florida) and others to infer the orbital properties of binary star systems using just the radial velocity “error” reported by Gaia EDR3. This week, I updated the group on this project, and (shockingly) I was excited to share some lessons learned when building the infrastructure for this pipeline. In particular, I’ve been using Snakemake (or, Snakémaké, right?) to orchestrate the pipeline and I’ve been finding it very useful.
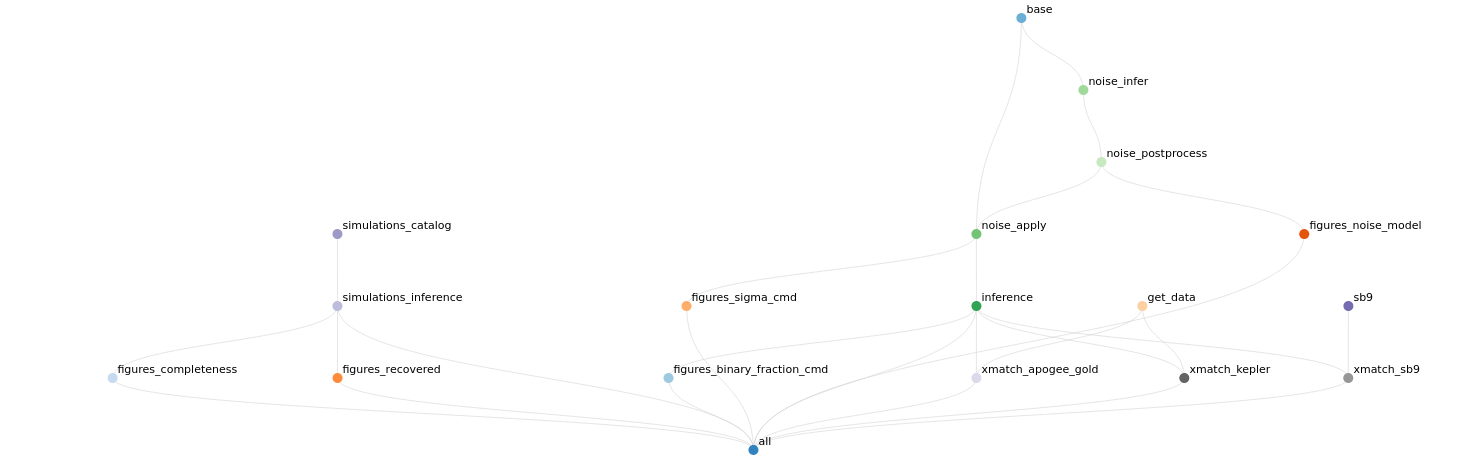
Snakemake is more or less an extremely glorified Makefile system (yeah ok I understand it’s more than that; don’t @ me) with lots of features that make it a good tool for scientific workflows in particular. I was originally drawn to it because it is more HPC cluster friendly than most of the other similar tools, but it also has good support for designing reproducible workflows. Today I shared a visualization of my pipeline workflow that was automatically generated by Snakemake. You can see a screenshot above and a rendered version of this interactive visualization here. This is fun (and useful!) because it shows the relationships between different scripts (nodes in the above network) that were executed to generate data products (represented by the connections between nodes). If you have scripts that generate figures, you can also include those in this visualization (try clicking on the sections in the sidebar).
The group asked about support for versioning workflows and including development versions of dependencies. The former is pretty well supported in some cases; for example, if you want to generate results for a range of different parameters for a particular script that is fairly straightforward. On the other hand, I’ve really struggled to construct a good workflow where one of the dependencies is a library that you’re developing in parallel. It might be possible, for example, to include installing your package as a node in your workflow, but I’ve found that that can end up causing you to re-run steps that were unnecessary.
There is some overhead to getting started with this toolset, but if you’re trying to orchestrate a set of analysis scripts with interdependencies, I think that Snakemake could be a good fit. The code for this project is (naturally) on GitHub at one-datum/pipeline so feel free to check it out!